# Ontop with Dremio
In this tutorial we present step-by-step the way of connecting Dremio to Ontop. We show how to integrate uni1 data saved in the PostgreSQL database and uni2 data saved in plain JSON files into one Dremio data space.
We provide a Docker-compose script, which is both launching Dremio and the PostgreSQL database. Go at the root of the ontop-tutorial git repository and run the following commands:
cd federation/dremio
docker-compose pull && docker-compose up --build
Once Dremio is initialized, visit http://localhost:9047 (opens new window). Before you continue, we recommend you to see the following tutorials provided by Dremio:
By following the instructions in Working with your first dataset (opens new window), we create a space named university as shown below.
Add and save the new data space:
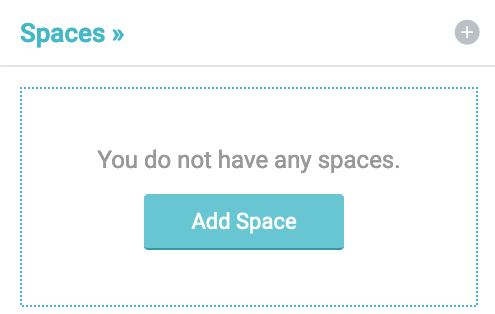
It will be our data space in which we integrate data from various sources.
The uni1 data is contained in a PostgreSQL database named university-session1. We are ready to add our database as a new datasource into Dremio:
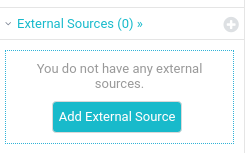
Select PostgreSQL:
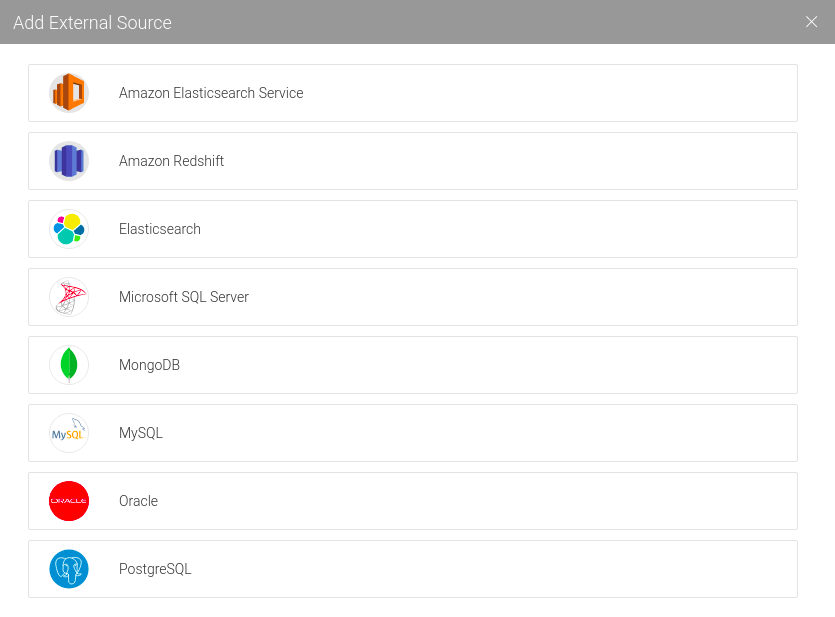
Enter the required JDBC information (password: postgres):
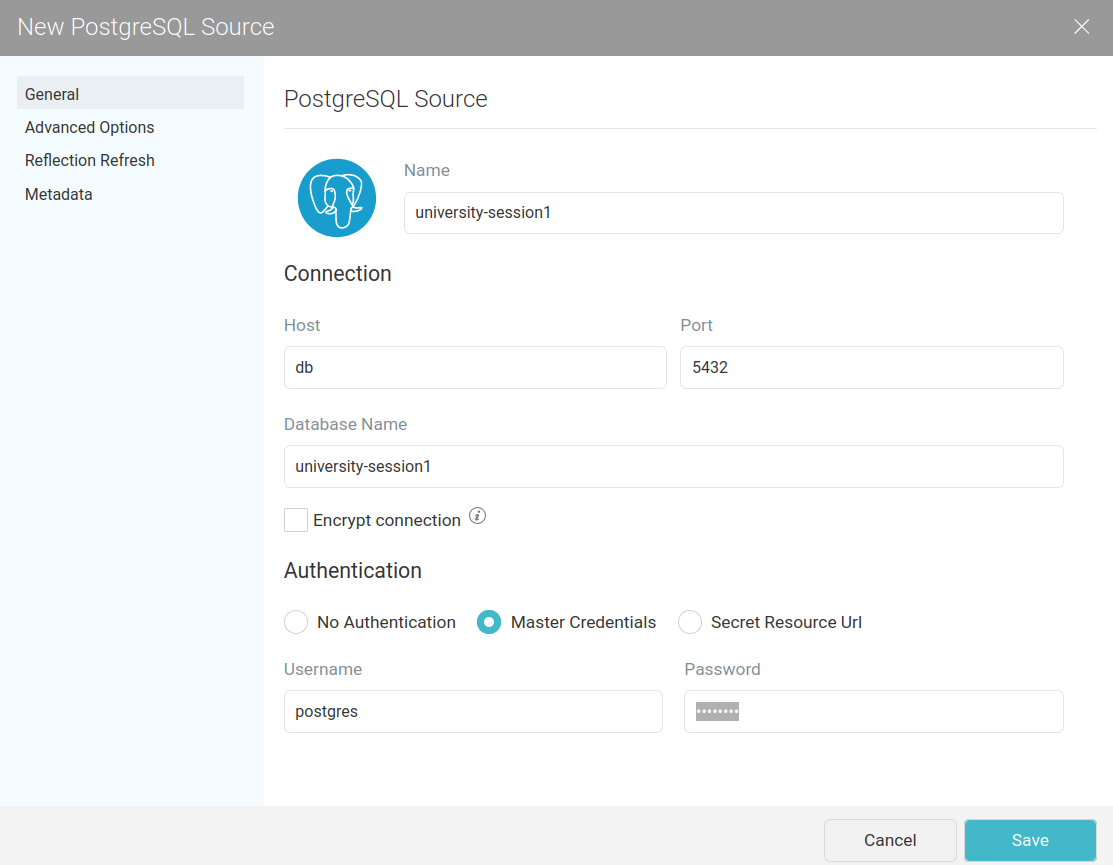
Now we see the tables in university-session1:
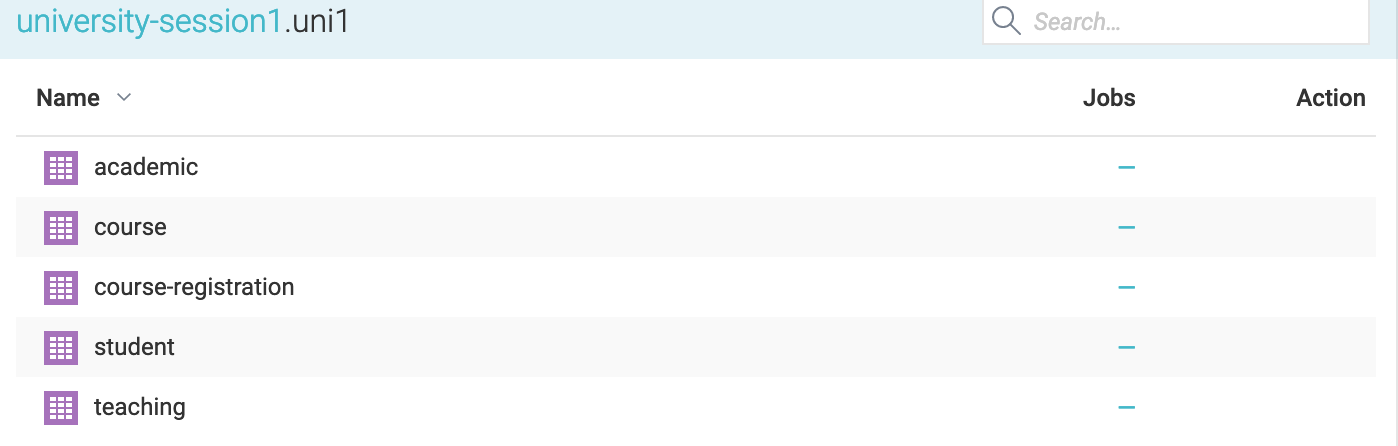
We add the table course-registration as dataset into university. We rename it to uni1-registration.
Save the other datasets in similar manner.
Now we add the uni2 data from here as a JSON data source:
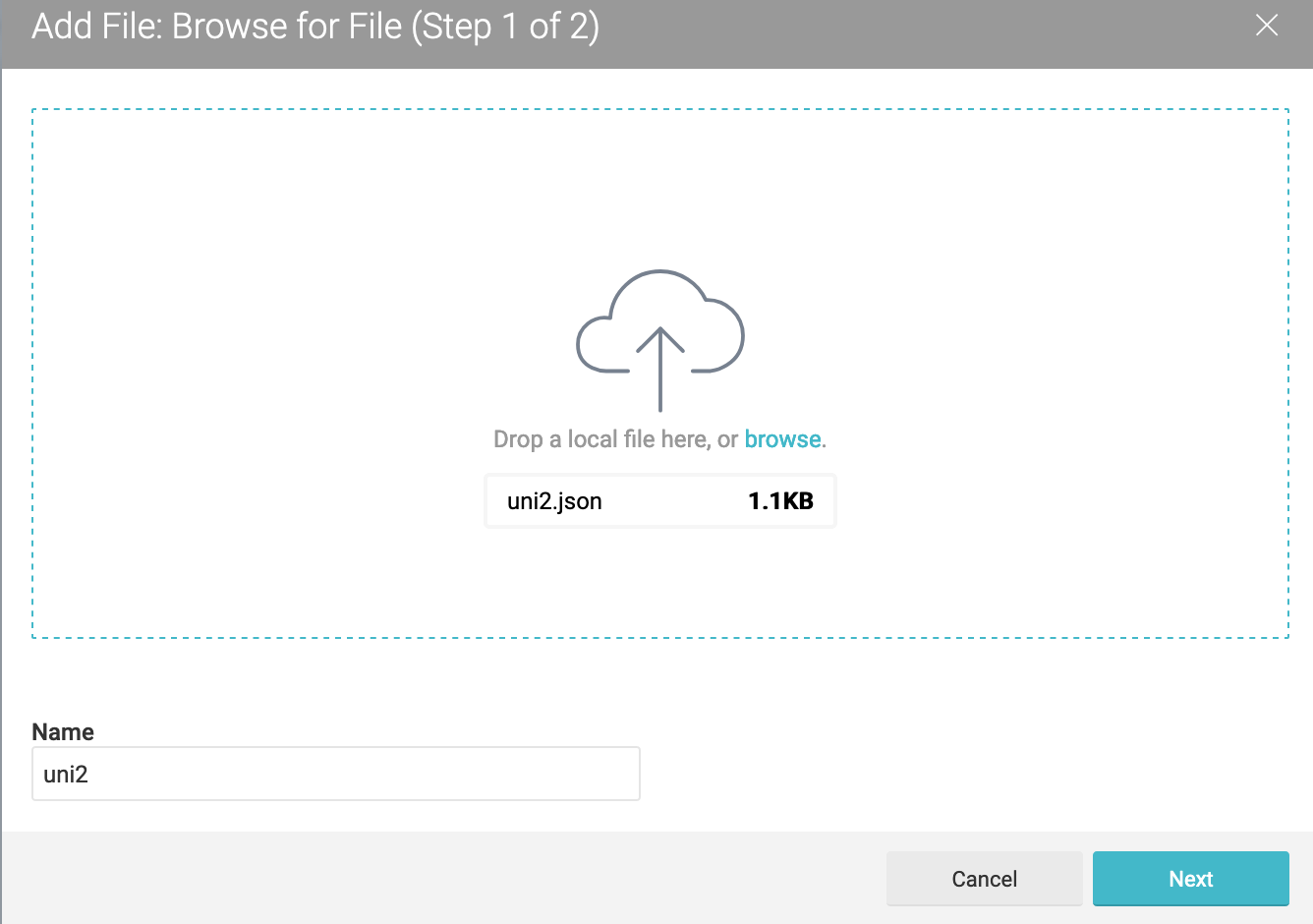
The uni2 JSON data can be seen as follows:
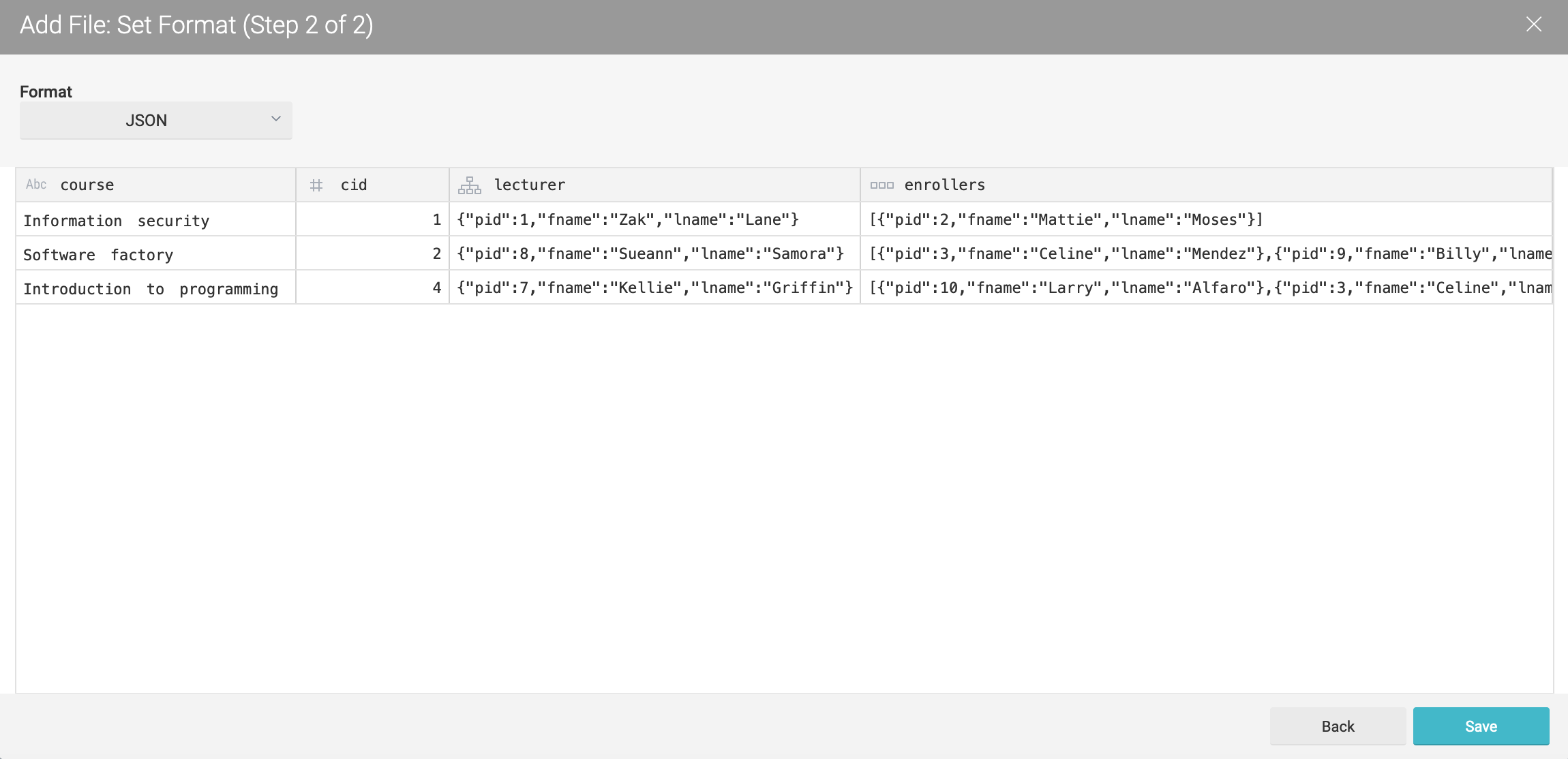
JSON files usually contain nested data. However, Ontop cannot directly query nested data. For this reason, in order to make our JSON data queryable by Ontop, first we need to extract relevant group of elements, and save these groups as datasets.
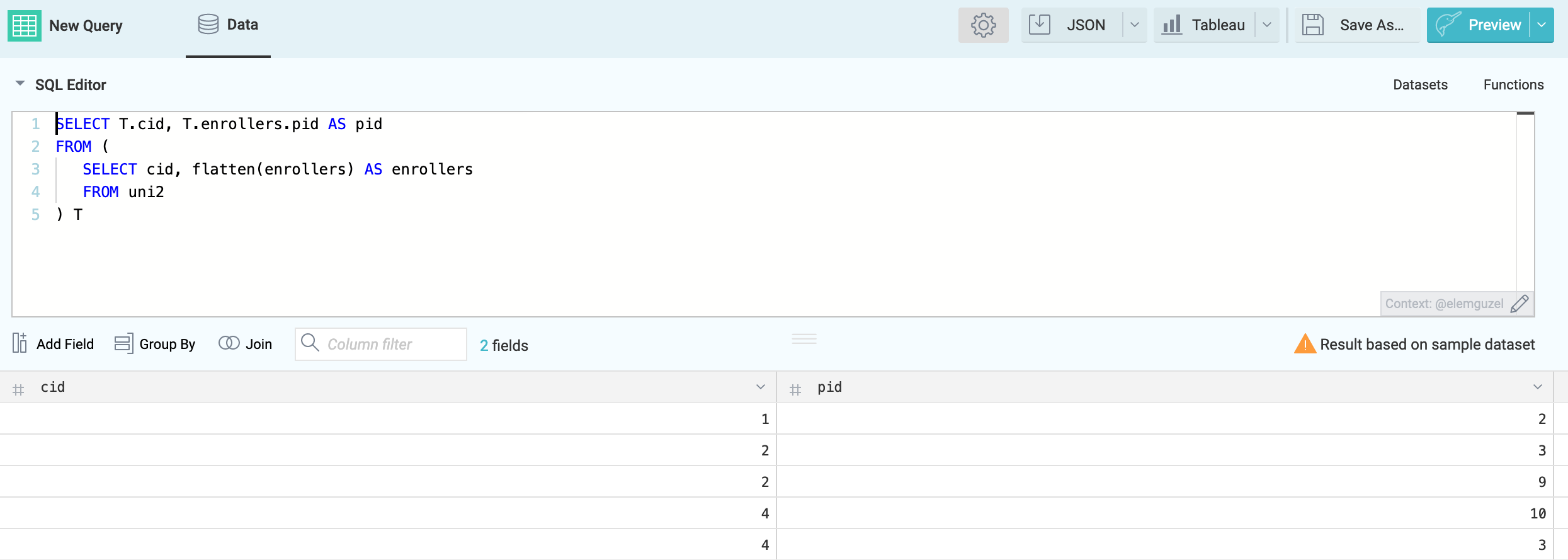
With the following SQL query we create an uni2-registration dataset and save it into the data space university:
SELECT T.cid, T.enrollers.pid AS pid
FROM (
SELECT cid, flatten(enrollers) AS enrollers
FROM uni2
) T
The following SQL expression flattens the array of enrollers:
flatten(enrollers) AS enrollers
Save As the data set with the name uni2-registration.
Create a dataset named uni2-course in a similar manner:
SELECT course, cid FROM uni2
Create a uni2-student dataset with a bit more involved SQL that flattens the array of students:
SELECT T.enrollers."pid" AS pid,T.enrollers."fname" AS fname, T.enrollers."lname" AS lname
FROM(
SELECT flatten(enrollers) AS enrollers
FROM uni2
) T
Create uni2-teaching dataset with the following SQL:
SELECT cid, uni2.lecturer.pid AS pid
FROM uni2
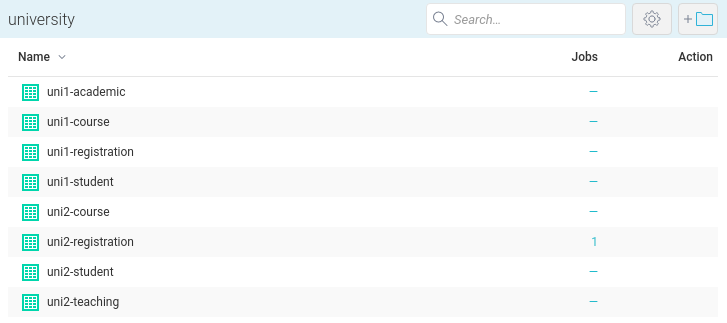
Now we can list all the datasets we saved in the university space:
Finally we are ready to connect Dremio to Ontop. Dremio can be connected to Ontop through its JDBC interface. By following the instructions provided in here (opens new window), we provide to Ontop the following JDBC connection information in a ".properties file" for a Dremio instance running on the localhost:
jdbc.url=jdbc\:dremio\:direct\=localhost\:31010
jdbc.driver=com.dremio.jdbc.Driver
jdbc.user=dremiotest
jdbc.password=dremiotest
Dremio JDBC driver can be downloaded from here (opens new window).
Over an OBDA setting containing the following mapping assertions:
[PrefixDeclaration]
: http://example.org/voc#
ex: http://example.org/
owl: http://www.w3.org/2002/07/owl#
rdf: http://www.w3.org/1999/02/22-rdf-syntax-ns#
xml: http://www.w3.org/XML/1998/namespace
xsd: http://www.w3.org/2001/XMLSchema#
foaf: http://xmlns.com/foaf/0.1/
obda: https://w3id.org/obda/vocabulary#
rdfs: http://www.w3.org/2000/01/rdf-schema#
[MappingDeclaration] @collection [[
mappingId uni1-student
target :uni1/student/{s_id} a :Student ; foaf:firstName {first_name}^^xsd:string ; foaf:lastName {last_name}^^xsd:string .
source SELECT * FROM "university"."uni1-student"
mappingId uni1-attends
target :uni1/student/{s_id} :attends :uni1/course/{c_id}/{title} .
source SELECT "uni1-registration".s_id, "uni1-course".c_id, "uni1-course".title FROM "university"."uni1-registration", "university"."uni1-course" WHERE "uni1-registration".c_id = "uni1-course".c_id
mappingId uni2-student
target :uni2/student/{pid} a :Student ; foaf:firstName {fname}^^xsd:string ; foaf:lastName {lname}^^xsd:string .
source SELECT * FROM "university"."uni2-student"
mappingId uni2-attends
target :uni2/student/{pid} :attends :uni2/course/{cid}/{course} .
source SELECT "uni2-registration".pid, "uni2-course".cid, "uni2-course".course FROM "university"."uni2-registration", "university"."uni2-course" WHERE "uni2-registration".cid = "uni2-course".cid
]]
Now we can execute the following SPARQL query on Ontop:
PREFIX : <http://example.org/voc#>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
SELECT ?course ?firstName ?lastName {
?student :attends ?course .
?student foaf:firstName ?firstName .
?student foaf:lastName ?lastName .
}